### Programming Experiences
- Python
- Go
- R
- JavaScript
- C#
### Bioinfomatics Experiences (PhD)
- Proteomics and glycoproteomics data pipeline development
- In-silico analysis pipeline for glycosylation evolution
- 5' UTR open reading frame database uPEPerroni development
- ...
### Recent projects (Postdoctoral)
- [Curtain](https://github.com/noatgnu/curtain) and [CurtainPTM](https://github.com/noatgnu/curtainptm), data analysis sharing tools
- [Copica](https://github.com/noatgnu/copica), protein copy number database
- Metabolomics data analysis pipeline
- [Adept](https://github.com/noatgnu/adeptpy), data analysis sharing tools and pipeline
- ...
### Curtain and CurtainPTM
- Shared Python backend
- Separated frontend written using JavaScript
- Each saved datasets are given a unique ids which could be used to access the data as well as the associated settings
```python
hash = ""
req = json_decode(self.request.body.decode("utf-8"))
if "password" in req:
if req["password"]:
hash = bcrypt.hashpw(req["password"].encode("utf-8"), bcrypt.gensalt())
uni = str(uuid4())
with open(f"files/{uni}.json", "wb") as data:
data.write(self.request.body)
f = File(password=hash, filename=uni)
```
### CurtainPTM Example
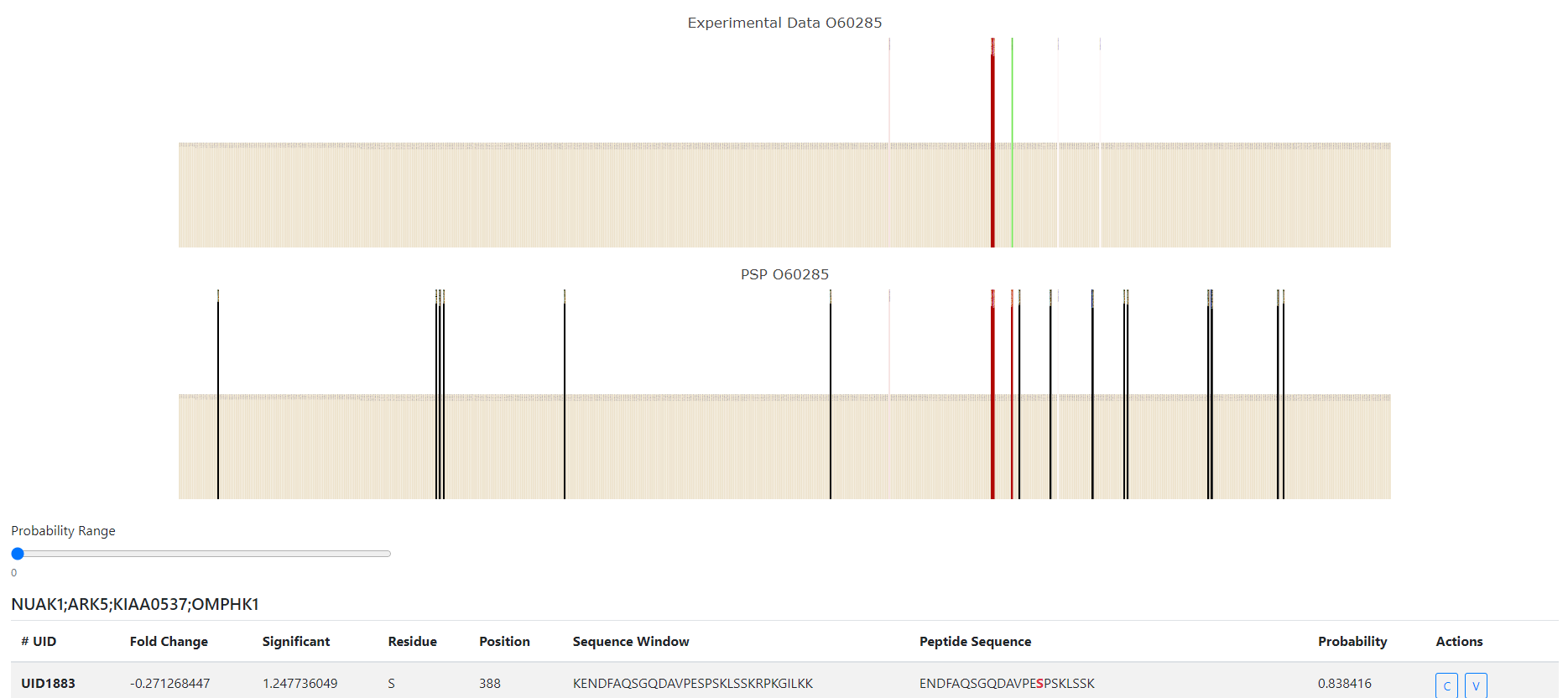
### Copica
- A copy number database with a focus on cell line used in Parkinson's research
- Included a JavaScript implementation of a protein copy number algorithm
```typescript
this.ruler = new ProteomicRuler(this.histoneDB, e.df, e.IntensityCols, e.IdentifierCol, e.MolecularMassCol, e.Ploidy)
const data: any[] = []
const columns = this.ruler.intensityCols
for (const r of this.ruler.df) {
if (r[e.IdentifierCol] !== "") {
for (let i = 0; i < columns.length; i++) {
data.push({
"Gene names": r[e.GeneNameCol],
"Accession IDs": r[e.IdentifierCol].toUpperCase(),
"Copy number": r[columns[i]+"_copyNumber"],
"Rank": r[columns[i]+"_copyNumber_rank"],
"Cell type": "UserCellType"+i, "Experiment type": "Experiment"+i, "Fraction": i, "Condition": "Standard", "label": "UserCellType"+i+"Standard"
})
}
}
}
this.scatterData = new DataFrame(data)
this.userData.updateData(this.scatterData)
this.finishedProcessing.next(true)
```
### Copica Example
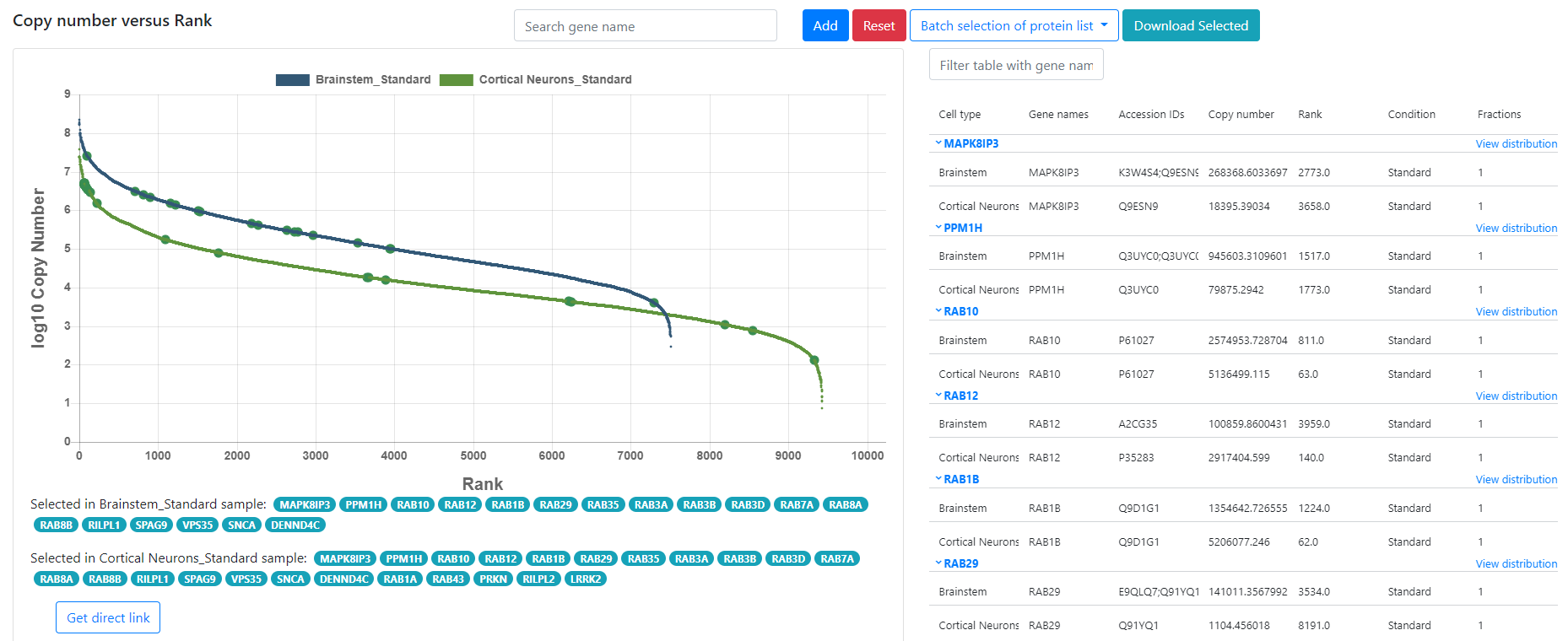
### Metabolomics data analysis pipeline
- Working with collaborators with effect of Parkinson's on different cell lines metabolomics profile.
- Use ExtraTreeClassifier to identify important features.
```python
def get_important(data, experiments):
etc = ExtraTreesClassifier()
d = data.current_df[experiments].T
x = d.values
d.reset_index(inplace=True)
y = []
for i in d[d.columns[0]]:
y.append(i.split(".")[0])
etc.fit(x, y)
extra = pd.Series(etc.feature_importances_, index=d.columns[1:])
return extra
```
### Adept
- Data statistical analysis tool
- Allow saving and sharing of your workflows
- Implementation of a special data structure that allow rewinding of the analysis
- Separate frontend and backend
```python
class Data:
def __init__(self, df=None, file_path=None, parent=None, operation=None, history=False, index=None):
def initiate_history(self):
self.history = [self]
def rewind(self, offset=1):
def forward(self, offset=1):
def statistical_analysis_methods(self):
```